New development of membrane protein structure analysis technology
 The breakthrough of a series of technical bottlenecks such as protein expression, dissolution and crystallization has made it possible to study the atomic structure of membrane proteins.
The breakthrough of a series of technical bottlenecks such as protein expression, dissolution and crystallization has made it possible to study the atomic structure of membrane proteins.
About 30% of the proteins in cells are membrane proteins, but people still don't know the atomic structure of these membrane proteins. So far, less than 1% of the data in the PDB (Protein Data Bank) structural database is the structural data of membrane proteins. This is not to say that the structure of membrane proteins is not important, on the contrary, membrane receptor proteins are very important, they are the target of most drugs. However, due to the lack of good soluble protein expression technology, there is not enough protein crystals for structural analysis. Even the subject of structural genomics, which aims to analyze the structure of each protein family, has not focused on membrane proteins because of technical difficulties.
Now, through the unremitting efforts of traditional structural biologists and structural genomics scientists, a series of technical bottlenecks in protein expression, dissolution and crystallization have been broken through, and some results have already appeared.
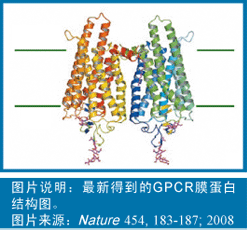
In 2007 and 2008, the academic community published several landmark articles in succession. These articles are all about the traditional structural difficulties of the G protein-coupled receptor (GPCR) crystal structure. Thesis.
Structural genomics research organizations such as the New York Consortium on Membrane Protein Structures (NYCOMPS) and Center for Structures of Membrane Proteins (CSMP) and several other international research organizations are working Work together to develop an efficient and standardized technical process to study membrane protein structure. So far, structural information on several membrane proteins has been obtained. At the same time, solid-state NMR technology, which does not require the use of protein crystals to study the structure, has also been rapidly developed, but the technology needs further improvement in terms of sensitivity and resolution.
It is not yet possible to obtain a "universal and omnipotent" technical process to analyze all protein structures, so other research methods for different protein structure analysis need to be developed. We believe that more methods for studying membrane protein structure will appear in the next few years, and more membrane protein structure data will be found in PDB.
Cat Beds,Cute Cat Beds,Heated Cat Beds,Pets At Home Cat Beds
Hebei siuno household supplies co.,ltd , https://www.ciaosleep.com